-
 Systematic Review: Fragile X Syndrome Across the Lifespan with a Focus on Genetics, Neurodevelopmental, Behavioral and Psychiatric Associations
Systematic Review: Fragile X Syndrome Across the Lifespan with a Focus on Genetics, Neurodevelopmental, Behavioral and Psychiatric Associations -
 The Impact of Klotho in Cancer: From Development and Progression to Therapeutic Potential
The Impact of Klotho in Cancer: From Development and Progression to Therapeutic Potential -
 Genomic Rewilding of Domestic Animals: The Role of Hybridization and Selection in Wolfdog Breeds
Genomic Rewilding of Domestic Animals: The Role of Hybridization and Selection in Wolfdog Breeds -
 Neuronal Network Activation Induced by Forniceal Deep Brain Stimulation in Mice
Neuronal Network Activation Induced by Forniceal Deep Brain Stimulation in Mice -
 Chemical Evolution of Life on Earth
Chemical Evolution of Life on Earth
Journal Description
Genes
Genes
is a peer-reviewed, open access journal of genetics and genomics published monthly online by MDPI. The Spanish Society for Biochemistry and Molecular Biology (SEBBM) is affiliated with Genes and their members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Genetics and Heredity) / CiteScore - Q2 (Genetics (clinical))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 14.9 days after submission; acceptance to publication is undertaken in 2.6 days (median values for papers published in this journal in the second half of 2024).
- Recognition of Reviewers: Reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
2.8 (2023);
5-Year Impact Factor:
3.3 (2023)
Latest Articles
The Complete Mitochondrial Genome of Petalocephala arcuata Cai Et Kuoh, 1992 (Hemiptera: Cicadellidae: Ledrinae: Petalocephalini) and Its Phylogenetic Implications
Genes 2025, 16(5), 567; https://doi.org/10.3390/genes16050567 (registering DOI) - 10 May 2025
Abstract
Background/Aims: Ledrinae comprises about 460 described species across five tribes and represents an early-branching, morphologically distinctive lineage of leafhoppers, yet its intra-subfamilial relationships remain ambiguous owing to limited mitogenomic sampling. Here, we sequence and annotate the complete mitochondrial genome of Petalocephala arcuata—only
[...] Read more.
Background/Aims: Ledrinae comprises about 460 described species across five tribes and represents an early-branching, morphologically distinctive lineage of leafhoppers, yet its intra-subfamilial relationships remain ambiguous owing to limited mitogenomic sampling. Here, we sequence and annotate the complete mitochondrial genome of Petalocephala arcuata—only the 18th Ledrinae mitogenome—to broaden taxon coverage within the genus and furnish critical molecular data for rigorously testing Ledrinae monophyly and refining tribal and genus level phylogenetic hypotheses. Methods: In this study, we sequenced and annotated the complete mitochondrial genome of P. arcuata via Illumina sequencing and de novo assembly, and reconstructed the phylogeny of 62 Cicadellidae species using maximum likelihood and Bayesian inference methods. Results: The 14,491 bp circular mitogenome of P. arcuata contains 37 genes with 77.4% A+T. All PCGs use ATN start codons except ND5 (TTG), and codon usage is A or U biased. Of 22 tRNAs, only trnS1 lacks a DHU arm, while the others adopt the canonical cloverleaf structure. Bayesian inference and maximum likelihood analyses produced broadly congruent topologies with mostly high nodal support, recovering Ledrinae as monophyletic and clustering all Petalocephala species into a well‑supported clade. Conclusions: In this study, we enriched the molecular resources for the genus Petalocephala by sequencing, annotating, and analyzing the complete mitochondrial genome of P. arcuata. Phylogenetic reconstructions based on these genomic data align closely with previous morphological diagnoses, further confirming the monophyly of the genus Petalocephala.
Full article
(This article belongs to the Special Issue Molecular Evolution, Mitochondrial Genomics and Mitochondrial Genome Expression in Animals: 2024–2025)
Open AccessReview
Natural Bioproducts with Epigenetic Properties for Treating Cardiovascular Disorders
by
Olaia Martínez-Iglesias, Vinogran Naidoo, Iván Carrera, Lola Corzo and Ramón Cacabelos
Genes 2025, 16(5), 566; https://doi.org/10.3390/genes16050566 (registering DOI) - 10 May 2025
Abstract
Cardiovascular disorders (CVDs) are the leading cause of mortality worldwide, highlighting an urgent need for innovative therapeutic strategies. Recent advancements highlight the potential of naturally derived bioproducts with epigenetic properties to offer protection against CVDs. These compounds act on key epigenetic mechanisms, DNA
[...] Read more.
Cardiovascular disorders (CVDs) are the leading cause of mortality worldwide, highlighting an urgent need for innovative therapeutic strategies. Recent advancements highlight the potential of naturally derived bioproducts with epigenetic properties to offer protection against CVDs. These compounds act on key epigenetic mechanisms, DNA methylation, histone modifications, and non-coding RNA regulation to modulate gene expression essential for cardiovascular health. This review explores the effects of various bioproducts, such as polyphenols, flavonoids, and other natural extracts, on these epigenetic modifications and their potential benefits in preventing and managing CVDs. We discuss recent discoveries and clinical applications, providing insights into the epigenetic regulatory mechanisms of these compounds as potential epidrugs, naturally derived agents with promising therapeutic prospects in epigenetic therapy for CVDs.
Full article
(This article belongs to the Section Epigenomics)
Open AccessArticle
Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea)
by
Ting Luo, Qianwen Zhang, Siyu Pang, Yanting Qin, Bin Zhang and Xun Bian
Genes 2025, 16(5), 565; https://doi.org/10.3390/genes16050565 (registering DOI) - 10 May 2025
Abstract
Background: Lonchodidae is the largest family within the order Phasmatodea, and although many studies have been conducted on this family, the monophyly of the family has not been established. Methods: Eight mitogenomes from Lonchodidae, including the first complete mitogenomes of four genera, were
[...] Read more.
Background: Lonchodidae is the largest family within the order Phasmatodea, and although many studies have been conducted on this family, the monophyly of the family has not been established. Methods: Eight mitogenomes from Lonchodidae, including the first complete mitogenomes of four genera, were sequenced and annotated to explore their features and phylogenetic relationships. Results: The total length ranged from 15,942–18,021 bp, and the mitogenome consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region (CR). atp8 had the highest A + T content in Lonchodidae, except for Neohirasea stephanus and Asceles clavatus, in which the highest A + T contents were detected in nad6. The phylogenetic trees were reconstructed via Bayesian inference (BI) and maximum likelihood (ML) based on the PCG123 and PCG12 datasets. As the phylogenetic trees show, Necrosciinae is recognized as monophyletic, but the monophyly of Lonchodinae has not been supported. Gene deletion and rearrangement have occurred mainly in Lonchodidae and Aschiphasmatidae. The most common reason for gene rearrangements was tandem duplication random loss (TDRL), but trnI of Stheneboea repudiosa inverted into the CR. In addition, genes within the same family or genus share related sequences and conserved gene blocks. Conclusions: we expanded the mitochondrial genomic data for this family, thereby establishing a foundational dataset for future studies.
Full article
(This article belongs to the Special Issue Molecular Evolution, Mitochondrial Genomics and Mitochondrial Genome Expression in Animals: 2024–2025)
►▼
Show Figures
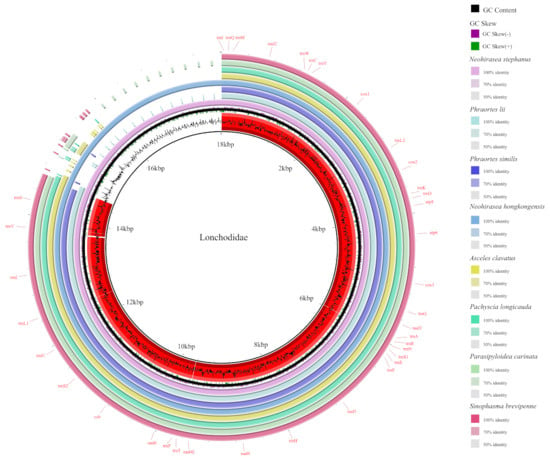
Figure 1
Open AccessArticle
Body Mass Index as an Example of a Negative Confounder: Evidence and Solutions
by
Zhu Liduzi Jiesisibieke and C. Mary Schooling
Genes 2025, 16(5), 564; https://doi.org/10.3390/genes16050564 (registering DOI) - 10 May 2025
Abstract
Background: Adequate control for confounding is key to many observational study designs. Confounders are often identified based on subject matter knowledge from empirical investigations. Negative confounders, which typically generate type 2 error, i.e., false nulls, can be elusive. Such confounders can be identified
[...] Read more.
Background: Adequate control for confounding is key to many observational study designs. Confounders are often identified based on subject matter knowledge from empirical investigations. Negative confounders, which typically generate type 2 error, i.e., false nulls, can be elusive. Such confounders can be identified comprehensively by using Mendelian randomization (MR) to search the wealth of publicly available data systematically. Here, to demonstrate the concept, we examined whether a common positive confounder, body mass index (BMI), is also a negative confounder of any common physiological exposures on health outcomes, overall and specifically by sex. Methods: We used an MR study, based on the largest overall and sex-specific genome-wide association studies of BMI (i.e., from the Genetic Investigation of ANthropometric Traits and the UK Biobank) and of relevant exposures likely affected by BMI, to assess, overall and sex-specifically, whether BMI is a negative confounder potentially obscuring effects of harmful physiological exposures. Inverse variance weighting was the main method. We assessed sex differences using a z-test. Results: BMI was a potential negative confounder for apolipoprotein B and total testosterone in men, and for both sexes regarding low-density lipoprotein cholesterol, choline, linoleic acid, polyunsaturated fatty acids, and cholesterol. Conclusions: Using BMI as an illustrative example, we demonstrate that negative confounding is an easily overlooked bias. Given negative confounding is not always obvious or known, using MR systematically to identify potential negative confounders in relevant studies may be helpful.
Full article
(This article belongs to the Section Human Genomics and Genetic Diseases)
►▼
Show Figures
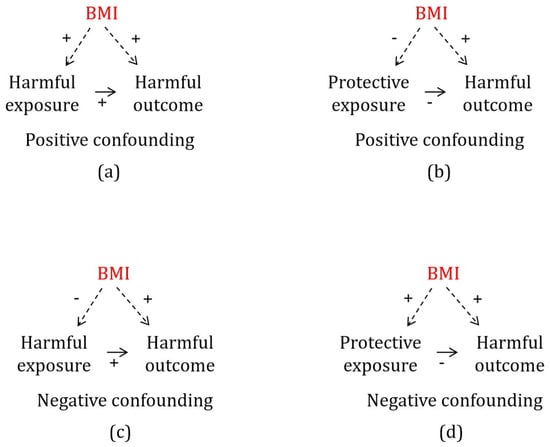
Figure 1
Open AccessArticle
FST-Based Marker Prioritization Within Quantitative Trait Loci Regions and Its Impact on Genomic Selection Accuracy: Insights from a Simulation Study with High-Density Marker Panels for Bovines
by
Sajjad Toghiani, Samuel E. Aggrey and Romdhane Rekaya
Genes 2025, 16(5), 563; https://doi.org/10.3390/genes16050563 (registering DOI) - 10 May 2025
Abstract
Background/Objectives: Genomic selection (GS) has improved accuracy compared to traditional methods. However, accuracy tends to plateau beyond a certain marker density. Prioritizing influential SNPs could further enhance the accuracy of GS. The fixation index (FST) allows for the identification of SNPs
[...] Read more.
Background/Objectives: Genomic selection (GS) has improved accuracy compared to traditional methods. However, accuracy tends to plateau beyond a certain marker density. Prioritizing influential SNPs could further enhance the accuracy of GS. The fixation index (FST) allows for the identification of SNPs under selection pressure. Although the FST method was shown to be able to prioritize SNPs across the whole genome and to increase accuracy, its performance could be further improved by focusing on the prioritization process within QTL regions. Methods: A trait with heritability of 0.1 and 0.4 was generated under different simulation scenarios (number of QTL, size of SNP windows around QTL, and number of selected SNPs within a QTL region). In total, six simulation scenarios were analyzed. Each scenario was replicated five times. The population comprised 30K animals from the last 2 generations (G9–G10) of a 10-generation (G1–G10) selection process. All animals in G9–10 were genotyped with a 600K SNP panel. FST scores were calculated for all 600K SNPs. Two prioritization scenarios were used: (1) selecting the top 1% SNPs with the highest FST scores, and (2) selecting a predetermined number of SNPs within each QTL window. GS accuracy was evaluated using the correlation between true and estimated breeding values for 5000 randomly selected animals from G10. Results: Prioritizing SNPs using FST scores within QTL window regions increased accuracy by 5 to 18%, with the 50-SNP windows showing the best performance. Conclusions: The increase in GS accuracy warrants the testing of the algorithm when the number and position of QTL are unknown.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
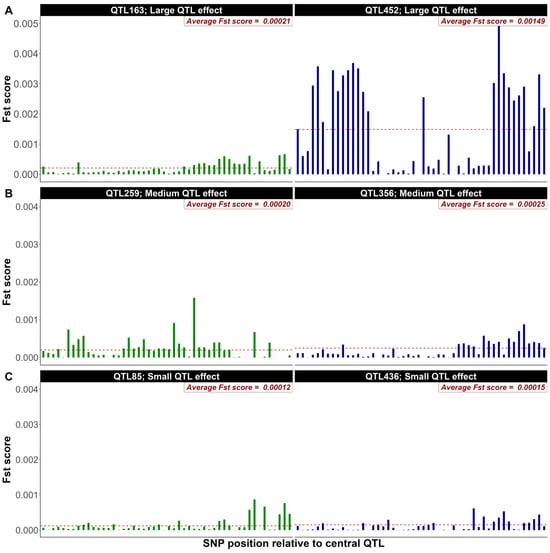
Figure 1
Open AccessArticle
Enhanced Reliability of the Evaluation of Fertility Traits in Pura Raza Española Horses Using Single-Step Genomic Best Linear Unbiased Prediction
by
Chiraz Ziadi, Mercedes Valera, Nora Laseca, Davinia Perdomo-González, Sebastián Demyda-Peyrás, Arancha Rodríguez-Sainz de los Terreros and Antonio Molina
Genes 2025, 16(5), 562; https://doi.org/10.3390/genes16050562 - 9 May 2025
Abstract
Background/Objectives: By simultaneously integrating both genotyped and non-genotyped animals into genetic evaluation, the single-step genomic BLUP method enhanced the accuracy of genetic assessments. This study aimed to compare the increase in prediction reliability (R2) between restricted maximum likelihood (REML) and single-step
[...] Read more.
Background/Objectives: By simultaneously integrating both genotyped and non-genotyped animals into genetic evaluation, the single-step genomic BLUP method enhanced the accuracy of genetic assessments. This study aimed to compare the increase in prediction reliability (R2) between restricted maximum likelihood (REML) and single-step genomic REML (ssGREML) in the Pura Raza Española (PRE) horse breed. Methods: The dataset comprised reproductive records for seven fertility traits from 47,502 females, with a total of 57,316 animals represented in the pedigree. A total of 4009 animals were genotyped using the EQUIGENE 90K SNP array, and 71,322 SNPs were retained for analysis after quality control. Genetic parameters were estimated using a multivariate model with the BLUPF90+ v2.60 software. Results: Heritability estimates were similar between REML and ssGREML, ranging from 0.07 for IF12 to 0.349 for ALF. An increase in R2 was observed with ssGREML compared to REML across all traits, with overall gains ranging from 2.20% to 3.71%. Among genotyped animals, R2 values ranged from 17.81% to 24.04%, while significantly lower values (0.80% to 2.34%) were observed in non-genotyped animals. Notably, individuals with low initial R2 values under the REML approach exhibited the most significant gains using ssGREML. This improvement was particularly pronounced among stallions with fewer than 40 controlled foals. Conclusions: Our results demonstrated that incorporating genomic data improves the reliability of genetic evaluations for mare fertility in PRE horses.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
Open AccessArticle
Mitochondrial Genomes of the Robberflies Clephydroneura jiangxiensis and Maira xizangensis (Diptera: Asilidae) and Phylogeny of Three Superfamilies
by
Keyao Zhang, Junhui Lu and Sheng-Quan Xu
Genes 2025, 16(5), 561; https://doi.org/10.3390/genes16050561 - 8 May 2025
Abstract
Background: Asilomorpha, an infraorder of predatory Diptera (Brachycera), is of significant evolutionary interest due to their remarkable ecological diversity, broad size range, and specialized feeding behaviors. However, phylogenetic studies of this group have been limited by sampling challenges. Methods: In this study, we
[...] Read more.
Background: Asilomorpha, an infraorder of predatory Diptera (Brachycera), is of significant evolutionary interest due to their remarkable ecological diversity, broad size range, and specialized feeding behaviors. However, phylogenetic studies of this group have been limited by sampling challenges. Methods: In this study, we sequenced the complete mitochondrial genomes of two Chinese endemic species, Clephydroneura jiangxiensis (C. jiangxiensis) and Maira xizangensis (M. xizangensis), using whole-genome random sequencing. By integrating these novel data with published sequences from NCBI, we reconstructed the phylogeny of three superfamilies (Asiloidea, Empidoidea, and Nemestrinoidea). Results: Both mitochondrial genomes exhibit the typical 37 genes (13 protein-coding genes, 22 tRNAs, and 2 rRNAs) and display pronounced AT bias. Congruent results from maximum likelihood analysis and Bayesian inference strongly supported the ideas that both new species are placed in Asilidae and that the Asilidae family is monophyletic. However, relationships among the three superfamilies remain unclear. Our results suggest that (1) although Asiloidea and Nemestrinidea are closely related, the potential positioning of Nemestrinoidea as an independent superfamily is worth investigating; and (2) Empidoidea may form a sister group to Asiloidea + Nemestrinidae, though this hypothesis requires further corroboration given the basal position of Hemipenthes hebeiensis (Bombyliidae). Conclusions: These findings highlight the need for expanded taxon sampling, particularly of underrepresented families, to resolve deep-level relationships within Asilomorpha. Clarifying the phylogenetic relationships within Asilomorpha will facilitate future investigations into their evolutionary origins and the evolution of characteristic traits.
Full article
(This article belongs to the Special Issue Molecular Evolution, Mitochondrial Genomics and Mitochondrial Genome Expression in Animals: 2024–2025)
►▼
Show Figures

Figure 1
Open AccessReview
The Role of Artificial Intelligence in Identifying NF1 Gene Variants and Improving Diagnosis
by
Vasiliki Sofia Grech, Kleomenis Lotsaris, Theano Eirini Touma, Vassiliki Kefala and Efstathios Rallis
Genes 2025, 16(5), 560; https://doi.org/10.3390/genes16050560 - 7 May 2025
Abstract
Neurofibromatosis type 1 (NF1) is an autosomal dominant disorder caused by mutations in the NF1 gene, typically diagnosed during early childhood and characterized by significant phenotypic heterogeneity. Despite advancements in next-generation sequencing (NGS), the diagnostic process remains challenging due to the gene’s complexity,
[...] Read more.
Neurofibromatosis type 1 (NF1) is an autosomal dominant disorder caused by mutations in the NF1 gene, typically diagnosed during early childhood and characterized by significant phenotypic heterogeneity. Despite advancements in next-generation sequencing (NGS), the diagnostic process remains challenging due to the gene’s complexity, high mutational burden, and frequent identification of variants of uncertain significance (VUS). This review explores the emerging role of artificial intelligence (AI) in enhancing NF1 variant detection, classification, and interpretation. A systematic literature search was conducted across PubMed, IEEE Xplore, Google Scholar, and ResearchGate to identify recent studies applying AI technologies to NF1 genetic analysis, focusing on variant interpretation, structural modeling, tumor classification, and therapeutic prediction. The review highlights the application of AI-based tools such as VEST3, REVEL, ClinPred, and NF1-specific models like DITTO and RENOVO-NF1, which have demonstrated improved accuracy in classifying missense variants and reclassifying VUS. Structural modeling platforms like AlphaFold contribute further insights into the impact of NF1 mutations on neurofibromin structure and function. In addition, deep learning models, such as LTC neural networks, support tumor classification and therapeutic outcome prediction, particularly in NF1-associated complications like congenital pseudarthrosis of the tibia (CPT). The integration of AI methodologies offers substantial potential to improve diagnostic precision, enable early intervention, and support personalized medicine approaches. However, key challenges remain, including algorithmic bias, limited data diversity, and the need for functional validation. Ongoing refinement and clinical validation of these tools are essential to ensure their effective implementation and equitable use in NF1 diagnostics.
Full article
(This article belongs to the Section Bioinformatics)
►▼
Show Figures
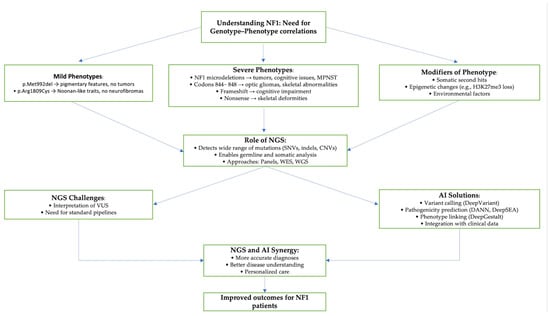
Figure 1
Open AccessArticle
Identification of Mustard Aldehyde Dehydrogenase (ALDH) Gene Family and Expression Analysis Under Salt and Drought Stress
by
Yuling Zheng, Shanshan Wang, Ling He, Rui Chen, Wei Zhang, Huachuan He, Hanbing Hu, Xiaoyun Liu, Heping Wan and Chunhong Wu
Genes 2025, 16(5), 559; https://doi.org/10.3390/genes16050559 - 7 May 2025
Abstract
Background/Objectives: Abiotic stresses severely constrain the yield of Brassica juncea, and aldehyde dehydrogenases (ALDHs) play a pivotal role in plant stress resistance. This study aims to systematically identify the ALDH gene family members in B. juncea and elucidate their expression patterns under salt
[...] Read more.
Background/Objectives: Abiotic stresses severely constrain the yield of Brassica juncea, and aldehyde dehydrogenases (ALDHs) play a pivotal role in plant stress resistance. This study aims to systematically identify the ALDH gene family members in B. juncea and elucidate their expression patterns under salt and drought stress. Methods: Using the Arabidopsis thaliana AtALDH proteins as seed sequences, BLASTp alignment was performed against the B. juncea whole-protein sequence database, combined with the conserved domain PF00171 of the ALDH proteins. A total of 39 BjALDH gene family members were identified, and their physicochemical properties, structures, phylogenetic relationships, interspecies collinearity, and intraspecies collinearity were analyzed. The qRT-PCR method was employed to quantify the relative expression levels of the BjALDH genes potentially associated with stress resistance under various treatments, and their effects on drought and salt stress tolerance were evaluated. Results: The results demonstrated that BjALDH were universally significantly upregulated under salt stress, while exhibiting predominantly upregulated trends under drought stress. These findings suggest that BjALDH may enhance plant resistance to both salt and drought stress by modulating the aldehyde metabolic pathways. Conclusions: This study provides a theoretical basis for elucidating the functional roles and molecular genetic mechanisms of the BjALDH gene family in B. juncea under salt and drought stress.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
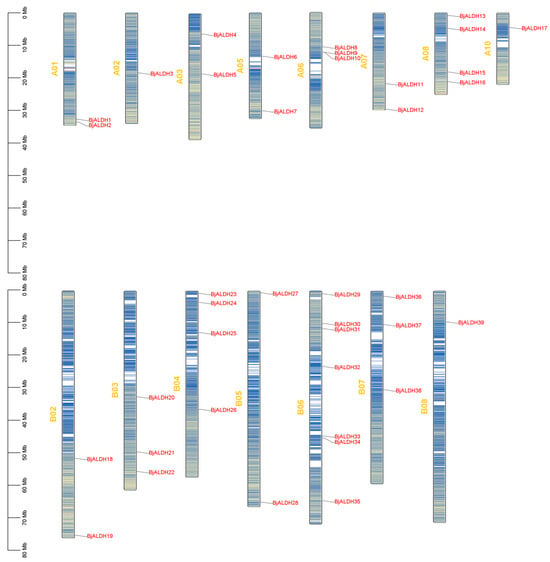
Figure 1
Open AccessArticle
De Novo Transcriptome Sequencing Analysis Revealed the Expression Patterns of Genes in Different Organs and the Molecular Basis of Polysaccharide Synthesis in Bletilla striata
by
Wenkui Liu, Jinxing Jiang, Zhonghai Tang, Zemao Yang and Jingping Qin
Genes 2025, 16(5), 558; https://doi.org/10.3390/genes16050558 - 6 May 2025
Abstract
Background: Bletilla striata (Thunb.) Rchb.f., a perennial medicinal plant in the genus Bletilla of the Orchidaceae family, is renowned for its hemostatic, anti-inflammatory, and tissue-regenerative properties. Despite the established importance of polysaccharides as key bioactive components in B. striata, the genes and
[...] Read more.
Background: Bletilla striata (Thunb.) Rchb.f., a perennial medicinal plant in the genus Bletilla of the Orchidaceae family, is renowned for its hemostatic, anti-inflammatory, and tissue-regenerative properties. Despite the established importance of polysaccharides as key bioactive components in B. striata, the genes and molecular mechanisms underlying their synthesis remain largely unexplored. Methods: This study conducted transcriptomic analysis on the roots, tubers, and leaves of B. striata, and identified gene expression profiles and candidate genes for polysaccharide synthesis in different organs. Results: The results indicated that there were 7699 differentially expressed genes (DEGs) in Tuber vs. Leaf, 7695 DEGs in Luber vs. Root, and 6151 DEGs in Tuber vs. Root. There were significant differences in polysaccharide metabolism pathways (photosynthesis, starch, and sucrose metabolism) in different organs of B. striata. The overall enrichment levels were ranked as tubers > leaves > roots. It is worth noting that enzyme genes involved in polysaccharide synthesis exhibit significant organ specificity, with HK genes expression significantly higher in roots than in tubers and leaves, PMM, GMPP, pgm, and UGP2 genes highly expressed in tubers, while scrK, manA, and GPI genes have similar expression levels in the three organs. Conclusions: These findings identify key enzyme genes involved in the synthesis of polysaccharides in B. striata, providing a theoretical framework for enhancing its medicinal value through genetic improvement.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessReview
Leveraging Spatial Transcriptomics to Decode Craniofacial Development
by
Jeremie Oliver Piña, Resmi Raju, Aye Chan Myo, Evan Stipano, Malachi Wright and Rena N. D’Souza
Genes 2025, 16(5), 557; https://doi.org/10.3390/genes16050557 (registering DOI) - 3 May 2025
Abstract
Understanding how intricate cellular networks and signaling pathways communicate during the formation of craniofacial tissues like the palate and tooth has been the subject of intense investigation for several decades. Both organ systems undergo patterning morphogenesis and the subsequent terminal differentiation of matrix-producing
[...] Read more.
Understanding how intricate cellular networks and signaling pathways communicate during the formation of craniofacial tissues like the palate and tooth has been the subject of intense investigation for several decades. Both organ systems undergo patterning morphogenesis and the subsequent terminal differentiation of matrix-producing cells that form biomineralized matrices like bone, enamel, dentin, and cementum. Until recently, gene expression profiles could only be assessed for a select number of cells without the context of the entire milieu of genes expressed by neighboring cells and tissues. Today, the cutting-edge field of spatial transcriptomics offers a remarkable suite of innovative technologies of multiplex gene analyses and imaging that can assess the expression of a vast library of genes that are present in situ during normal and abnormal conditions. In this review, we summarize some key technologies which have in recent years enabled an unprecedented breadth and depth of transcriptomic analyses in craniofacial development. We focus in detail on select methods that our research group has applied to better understand the cellular and molecular events that drive palate and tooth development. Our overall goal is to unravel the complexities of these unique biological systems to provide meaningful biological insights into the cellular and molecular events that drive normal development. As a work-in-progress, we strive for a deeper understanding of the temporal and spatial gene expression profiles within cells and tissues during normal and abnormal palate and tooth development. Such knowledge provides the framework for further studies that can characterize the function of new or novel genes that have the potential of serving as therapeutic targets for correcting disorders like cleft palate and tooth agenesis.
Full article
(This article belongs to the Special Issue Genetic, Epigenetic and Environmental Factors in Dental Development and Pathologies: Genes, Interactions and Dental Development)
►▼
Show Figures

Figure 1
Open AccessArticle
Clinicopathological Characteristics of Ovarian and Breast Cancer in PALB2, RAD51C, and RAD51D Germline Pathogenic Variant Carriers
by
Jella-Rike J. A. H. Spijkervet, L. Lanjouw, L. P. V. Berger, M. D. Dorrius, B. van der Vegt and G. H. de Bock
Genes 2025, 16(5), 556; https://doi.org/10.3390/genes16050556 - 2 May 2025
Abstract
Background/Objectives: Germline pathogenic variants (GPVs) in PALB2, RAD51C, and RAD51D increase breast cancer (BC) and ovarian cancer (OC) risk. Limited data on clinicopathological characteristics of BC and OC in women with these GPVs hamper guideline development. Therefore, this study aims to
[...] Read more.
Background/Objectives: Germline pathogenic variants (GPVs) in PALB2, RAD51C, and RAD51D increase breast cancer (BC) and ovarian cancer (OC) risk. Limited data on clinicopathological characteristics of BC and OC in women with these GPVs hamper guideline development. Therefore, this study aims to describe these characteristics in a consecutive series of female PALB2, RAD51C, and RAD51D GPV carriers. Methods: Women with a PALB2, RAD51C, or RAD51D GPV determined before July 2023 at the University Medical Center Groningen were included. Cancer diagnoses were obtained through linkage with the Dutch Nationwide Pathology Databank (Palga). Median onset age and histopathological subtypes were compared to the data of The Netherlands Cancer Registry (NCR). Results: Among 164 GPV carriers (125 PALB2, 30 RAD51C, and 9 RAD51D), 54 BC and 6 OC cases were identified. The median BC onset age was 52 (n = 50), 71 (n = 3), and 43 years (n = 1) for PALB2, RAD51C, and RAD51D, respectively, compared with 62 years in the NCR. No BC histological subtype differences were observed in PALB2 carriers. The populations of RAD51C and RAD51D carriers were too small to compare to NCR data. No OC cases occurred in PALB2 carriers. The median OC onset age was 66 (n = 4) and 56 years (n = 2) for RAD51C and RAD51D carriers, respectively, versus 67 years in the NCR. All RAD51D carriers had high-grade serous carcinoma, compared to 51.5% in the NCR. Conclusions: Differences in onset age and histological subtypes were observed between GPV carriers and national data. Further research on cancer characteristics is needed to optimize counseling and cancer prevention in these women.
Full article
(This article belongs to the Special Issue Genetics and Genomics of Human Breast Cancer)
►▼
Show Figures

Figure 1
Open AccessArticle
Genome-Wide Characterization and Analysis of the FH Gene Family in Medicago truncatula Under Abiotic Stresses
by
Jiatong Wang and Chunyang Zhou
Genes 2025, 16(5), 555; https://doi.org/10.3390/genes16050555 - 1 May 2025
Abstract
Background: The formin family proteins play an important role in guiding the assembly and nucleation of linear actin and can promote the formation of actin filaments independently of the Arp2/3 complex. As a key protein that regulates the cytoskeleton and cell morphological structure,
[...] Read more.
Background: The formin family proteins play an important role in guiding the assembly and nucleation of linear actin and can promote the formation of actin filaments independently of the Arp2/3 complex. As a key protein that regulates the cytoskeleton and cell morphological structure, the formin gene family has been widely studied in plants such as Arabidopsis thaliana and rice. Methods: In this study, we conducted comprehensive analyses, including phylogenetic tree construction, conserved motif identification, co-expression network analysis, and transcriptome data mining. Results: A total of 18 MtFH gene family members were identified, and the distribution of these genes on chromosomes was not uniform. The phylogenetic tree divided the FH proteins of the four species into two major subgroups (Clade I and Clade II). Notably, Medicago truncatula and soybean exhibited closer phylogenetic relationships. The analysis of cis-acting elements revealed the potential regulatory role of the MtFH gene in light response, hormone response, and stress response. GO enrichment analysis again demonstrated the importance of FH for reactions such as actin nucleation. Expression profiling revealed that MtFH genes displayed significant transcriptional responsiveness to cold, drought, and salt stress conditions. And there was a temporal complementary relationship between the expression of some genes under stress. The protein interaction network indicated an interaction relationship between MtFH protein and profilin, etc. In addition, 22 miRNAs were screened as potential regulators of the MtFH gene at the post-transcriptional level. Conclusions: In general, this study provides a basis for deepening the understanding of the physiological function of the MtFH gene and provides a reference gene for stress resistance breeding in agricultural production.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
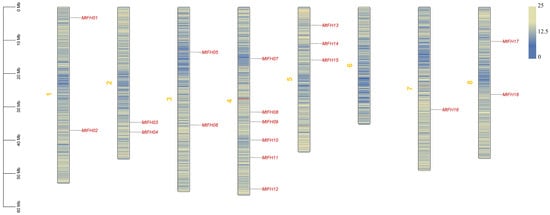
Figure 1
Open AccessArticle
The Complete Mitochondrial Genome of Red Costate Tiger Moth (Aloa lactinea [Cramer, 1777]), and Phylogenetic Analyses of the Subfamily Arctiinae
by
Chengrong Pan, Sheng Xu, Yu Shu and Jie Fang
Genes 2025, 16(5), 554; https://doi.org/10.3390/genes16050554 - 30 Apr 2025
Abstract
Background/Objectives: Aloa lactinea, class Insecta, order Lepidoptera, superfamily Noctuoidea, family Erebidae, and subfamily Arctiinae, is a polytrophic agricultural pest. However, there are still many sequences missing for Arctiinae from mitochondrial whole-genome sequences. Methods: In this study, we determined and analyzed the complete
[...] Read more.
Background/Objectives: Aloa lactinea, class Insecta, order Lepidoptera, superfamily Noctuoidea, family Erebidae, and subfamily Arctiinae, is a polytrophic agricultural pest. However, there are still many sequences missing for Arctiinae from mitochondrial whole-genome sequences. Methods: In this study, we determined and analyzed the complete mitochondrial genome sequence of A. lactinea. Furthermore, based on the sequencing results, we used the Bayesian inference, maximum likelihood, and maximum reduction methods to analyze the phylogenies of 18 species of the Hypophora subfamily. Results: The mitochondrial genome was found to be a circular double-stranded DNA with a length of 15,380 bp and included 13 protein-coding genes (PCGS), 22 tRNA genes, 2 rRNA genes, and one control region. With the exception of tRNASer(AGC), all the tRNA genes could form conventional clover structures. There were 23 intergenic spacer regions with lengths of 1–52 bp and six gene overlaps with lengths of 1–8 bp. The control region was located between rrnS and tRNAMet genes and comprised 303 bp and an AT content of 74.25%. Conclusions: The results showed that A. lactinea is closely related to Hyphantria cunea. Our results suggest that Syntomini is phylogenetically distinct from Arctiini and may warrant separate tribal status within Arctiinae. This study is dedicated to researching the mitochondrial genome and phylogenetic relationships of A. lactinea, providing a molecular basis for its classification.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
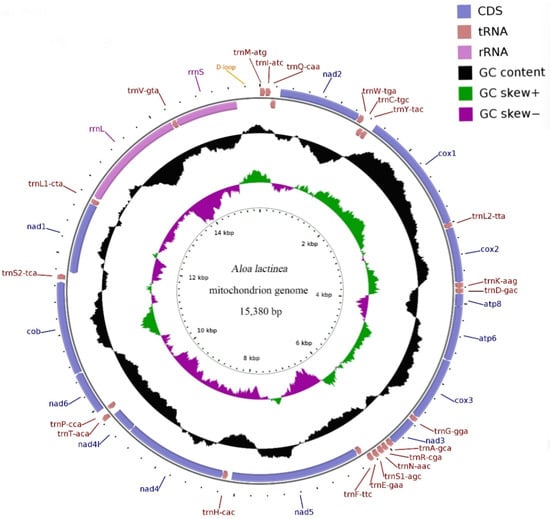
Figure 1
Open AccessReview
Updates on the Regulatory Framework of Edited Organisms in Brazil: A Molecular Revolution in Brazilian Agribusiness
by
Nicolau B. da Cunha, Jaim J. da Silva Junior, Amanda M. M. Araújo, Ludmila R. de Souza, Michel L. Leite, Gabriel da S. Medina, Gustavo R. Rodriguez, Renan M. dos Anjos, Júlio C. M. Rodrigues, Fabrício F. Costa, Simoni C. Dias, Elíbio L. Rech and Giovanni R. Vianna
Genes 2025, 16(5), 553; https://doi.org/10.3390/genes16050553 - 30 Apr 2025
Abstract
Genome editing technologies have revolutionized the production of microorganisms, plants, and animals with phenotypes of interest to agriculture. Editing previously sequenced genomes allows for the punctual, discreet, precise, and accurate alteration of DNA for genetic analysis, genotyping, and phenotyping, as well as the
[...] Read more.
Genome editing technologies have revolutionized the production of microorganisms, plants, and animals with phenotypes of interest to agriculture. Editing previously sequenced genomes allows for the punctual, discreet, precise, and accurate alteration of DNA for genetic analysis, genotyping, and phenotyping, as well as the production of edited organisms for academic and industrial purposes, among many other objectives. In this context, genome editing technologies have been causing a revolution in Brazilian agriculture. Thanks to the publication of Normative Resolution No. 16 (in Portuguese Resolução Normativa No. 16-RN16) in 2018, Brazilian regulatory authorities have adapted to the new genetic manipulation technologies available to the scientific community. This review aims to describe the effects of updates to the regulatory framework for edited organisms in Brazil and to point out their impacts on research and development of emerging technologies in the Brazilian agricultural sector. The implementation of RN16 rationalized the regulatory aspects regarding the production, manipulation, exploration and commercial release of edited organisms and led to the faster, cheaper and safer obtaining of edited technologies, which are more productive and better adapted to different environmental conditions in Brazil.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures
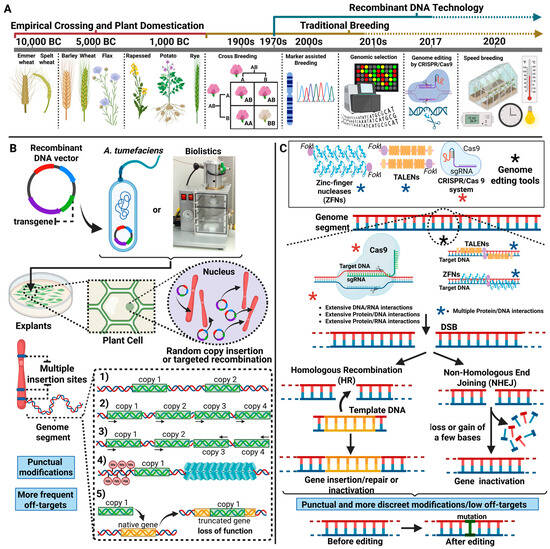
Figure 1
Open AccessReview
Non-Invasive Preimplantation Genetic Testing
by
Daniela N. Bakalova, Luis Navarro-Sánchez and Carmen Rubio
Genes 2025, 16(5), 552; https://doi.org/10.3390/genes16050552 - 30 Apr 2025
Abstract
To minimise the influence of chromosomal abnormalities during IVF treatment, embryos can be screened before transfer using preimplantation genetic testing. This typically involves an invasive trophectoderm biopsy at the blastocyst stage, where 4–8 cells are collected and analysed. However, emerging evidence indicates that,
[...] Read more.
To minimise the influence of chromosomal abnormalities during IVF treatment, embryos can be screened before transfer using preimplantation genetic testing. This typically involves an invasive trophectoderm biopsy at the blastocyst stage, where 4–8 cells are collected and analysed. However, emerging evidence indicates that, as embryos develop in vitro in culture media, they release cell-free DNA into the media, providing an alternative source of genetic material that can be accessed non-invasively. Spent blastocyst media samples that contain embryo cell-free DNA demonstrate high informativity rates and ploidy concordance when compared with the corresponding trophectoderm, inner cell mass, or whole blastocyst results. However, optimising this non-invasive approach requires several changes to embryo culture protocols, including additional embryo washes to tackle contamination and extending embryo culture time to maximise the amount of cell-free DNA released into the culture media. In this review, we discuss this novel non-invasive approach for aneuploidy detection and embryo prioritisation, as well as the current data and future prospects for utilising cell-free DNA analysis to identify structural rearrangements and single gene disorders.
Full article
(This article belongs to the Special Issue Current Advances and Future Perspectives on Preimplantation Genetic Testing)
►▼
Show Figures
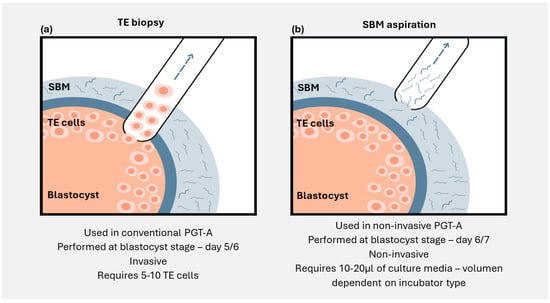
Figure 1
Open AccessArticle
Isolation, Enrichment and Analysis of Aerobic, Anaerobic, Pathogen-Free and Non-Resistant Cellulose-Degrading Microbial Populations from Methanogenic Bioreactor
by
Lyudmila Dimitrova, Yana Ilieva, Dilnora Gouliamova, Vesselin Kussovski, Venelin Hubenov, Yordan Georgiev, Tsveta Bratanova, Mila Kaleva, Maya M. Zaharieva and Hristo Najdenski
Genes 2025, 16(5), 551; https://doi.org/10.3390/genes16050551 - 30 Apr 2025
Abstract
Background: Nowadays, the microbial degradation of cellulose represents a new perspective for reducing cellulose waste from industry and households and at the same time obtaining energy sources. Methods: We isolated and enriched two aerobic (at 37 °C and 50 °C) and one anaerobic
[...] Read more.
Background: Nowadays, the microbial degradation of cellulose represents a new perspective for reducing cellulose waste from industry and households and at the same time obtaining energy sources. Methods: We isolated and enriched two aerobic (at 37 °C and 50 °C) and one anaerobic microbial consortium from an anaerobic bioreactor for biogas production by continuous subculturing on peptone cellulose solution (PCS) medium supplemented with 0.3% treated or untreated Whatman filter paper under static conditions. Samples were taken every 7 days until day 21 to determine the percentage of cellulose biodegradation. We determined the antimicrobial resistance of aerobic and anaerobic consortia and some single colonies by disc diffusion method, against 42 clinically applied antibiotics. PCR analyses were performed to search for the presence of eight genes for cellulolytic activity and nine genes for antibiotic resistance. By metagenomics analysis, the bacterial and fungal genus distributions in the studied populations were determined. Results: Aerobes cultured at 50 °C degraded cellulose to the greatest extent (47%), followed by anaerobes (24–38%) and aerobes (8%) cultured at 37 °C. The bacterial sequence analysis showed that the dominant phyla are Bacillota and Bacteroidetes and genera—Paraclostridium, Defluvitalea, Anaerobacillus, Acetivibrio, Lysinibacillus, Paenibacillus, Romboutsia, Terrisporobacter, Clostridium, Sporanaerobacter, Lentimicrobium, etc. in a different ratio depending on the cultivation conditions and the stage of the process. Some of these representatives are cellulolytic and hemicellulolytic microorganisms. We performed lyophilization and proved that it is suitable for long-term storage of the most active consortium, which degrades even after the 10th re-inoculation for a period of one year. We proved the presence of ssrA, ssrA BS and blaTEM genes. Conclusions: Our findings demonstrated the potential utility of the microbial consortium of anaerobes in the degradation of waste lignocellulose biomass.
Full article
(This article belongs to the Section Genes & Environments)
►▼
Show Figures
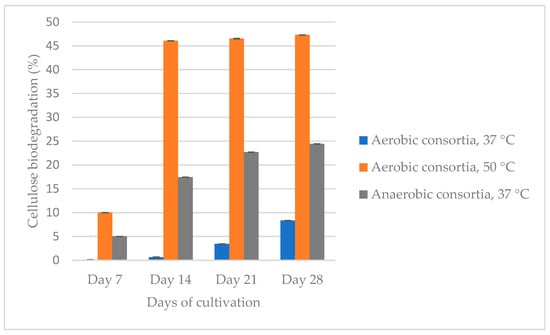
Figure 1
Open AccessArticle
Statistical Analysis of Reproductive Traits in Jinwu Pig and Identification of Genome-Wide Association Loci
by
Wenduo Chen, Ayong Zhao, Jianzhi Pan, Kai Tan, Zhiwei Zhu, Liang Zhang, Fuxian Yu, Renhu Liu, Liepeng Zhong and Jing Huang
Genes 2025, 16(5), 550; https://doi.org/10.3390/genes16050550 - 30 Apr 2025
Abstract
Background: The Jinwu pig is a novel breed created by crossbreeding Jinhua and Duroc pigs, displaying superior meat quality, strong adaptability to coarse feed, high production performance, and a rapid growth rate. However, research on its reproductive traits and genomic characteristics has not
[...] Read more.
Background: The Jinwu pig is a novel breed created by crossbreeding Jinhua and Duroc pigs, displaying superior meat quality, strong adaptability to coarse feed, high production performance, and a rapid growth rate. However, research on its reproductive traits and genomic characteristics has not been systematically reported. Methods: In this study, we investigated the genetic basis of reproductive traits in Jinwu pigs us-ing a genome-wide association study. We analyzed 2831 breeding records from 516 Jinwu sows to evaluate the effects of fixed factors (farrowing season, parity, and mated boar) on six reproductive traits: the total number of births (TNB), number born alive (NBA), number of healthy offspring produced (NHOP), weak litter size (WLS), number of stillbirths (NS), and number of mummies (NM). Results: A total of 771 genome-wide significant single-nucleotide polymorphisms (SNPs) and ten potential candidate genes associated with pig reproductive traits were identified: VOPP1, PGAM2, TNS3, LRFN5, ORC1, CC2D1B, ZFYYE9, TUT4, DCN, and FEZF1. TT-genotype-carrier individuals of the pleiotropic SNP rs326174997 exhibited significantly higher TNB, NBA, and NHOP trait-related phenotypic values. Conclusions: These findings provide a foundation for the reproductive breeding of Jinwu pigs and offer new insights into molecular genetic breeding in pigs.
Full article
(This article belongs to the Special Issue Advances in Pig Genetic and Genomic Breeding)
►▼
Show Figures
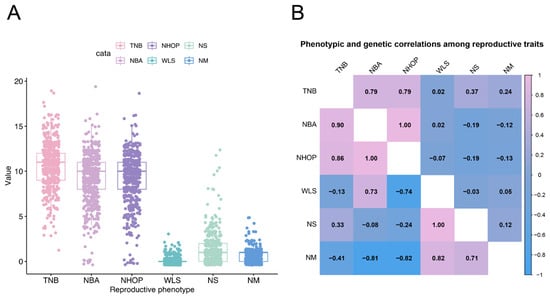
Figure 1
Open AccessArticle
Establishing Embryogenic Tissue Culture Workflow for Pineapple Cultivar 73–50
by
Ming Cheng, Yuri Trusov, Guoquan Liu, Yanfei Mao and Jose Ramon Botella
Genes 2025, 16(5), 549; https://doi.org/10.3390/genes16050549 - 30 Apr 2025
Abstract
Background: The development of an efficient tissue culture system is essential for advancing genetic transformation and genome editing in commercially important pineapple cultivars. However, a robust tissue culture workflow for the elite pineapple cultivar 73–50, enabling reliable transformation and plant regeneration is
[...] Read more.
Background: The development of an efficient tissue culture system is essential for advancing genetic transformation and genome editing in commercially important pineapple cultivars. However, a robust tissue culture workflow for the elite pineapple cultivar 73–50, enabling reliable transformation and plant regeneration is not established. Methods: A comparative analysis of hormone combinations, including 6-benzylaminopurine (BAP), α-naphthaleneacetic acid (NAA), picloram, and abscisic acid (ABA) was conducted. Transformation competence of 73–50 callus was tested using the iGUS reporter gene. Results: We established that 1 mg/L picloram and 0.5 µg/L ABA was the most effective combination for inducing friable embryogenic callus (FEC). FEC, composed of small, loosely associated cell clusters, is highly suitable for transformation but prone to browning during long-term culture. We optimized the conditions to minimize browning and support prolonged maintenance using a medium supplemented with 5 mg/L NAA. Transformation efficiency was demonstrated using the iGUS reporter gene, showing that FEC can be effectively transformed via both biolistic and Agrobacterium-mediated methods. For shoot regeneration, the optimal medium was found to contain 2 mg/L BAP. To standardize the assessment of callus development, we introduce a classification system describing distinct developmental stages. Conclusions: A detailed step-by-step protocol optimized for 73–50 cultivar facilitates efficient genetic improvement in pineapple, supporting both conventional transformation and DNA-free genome editing approaches.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
Phylogenomic and Evolutionary Insights into Lipoprotein Lipase (LPL) Genes in Tambaqui: Gene Duplication, Tissue-Specific Expression and Physiological Implications
by
Rômulo Veiga Paixão, Izabel Correa Bandeira, Vanessa Ribeiro Reis, Gilvan Ferreira da Silva, Fernanda Loureiro de Almeida O’Sullivan and Eduardo Sousa Varela
Genes 2025, 16(5), 548; https://doi.org/10.3390/genes16050548 - 30 Apr 2025
Abstract
Background/Objectives: Lipoprotein lipase (LPL) is a key enzyme in lipid metabolism, crucial for the hydrolysis of triglycerides in lipoproteins and maintaining lipid homeostasis in vertebrates. This study aims to characterize the lipoprotein lipase genes in the tambaqui (Colossoma macropomum) genome, investigating
[...] Read more.
Background/Objectives: Lipoprotein lipase (LPL) is a key enzyme in lipid metabolism, crucial for the hydrolysis of triglycerides in lipoproteins and maintaining lipid homeostasis in vertebrates. This study aims to characterize the lipoprotein lipase genes in the tambaqui (Colossoma macropomum) genome, investigating their evolutionary history from a phylogenomic perspective. Methods: Phylogenetic and syntenic analyses were used to identify the lpl gene copies in the tambaqui genome and expression patterns were examined across different tissues. A comparative analysis with lpl genes from other vertebrates was also conducted to assess evolutionary relationships and functional diversification. Results: We identified three lpl gene copies in the tambaqui genome: lpl1a, lpl1b, and the lesser-known member of the lipoprotein lipase subfamily, lpl2a. These proteins possess conserved sites essential for lipoprotein lipase function, with variations that may affect their physicochemical properties and lipolytic activity. Key amino acid variations, such as in the lid region and glycosylation sites, were observed among orthologs. Gene expression analysis showed high lpl1a and lpl2a expression in the liver, and lpl1b expression in the gonads, suggesting tissue-specific roles. Comparative analysis revealed distinct expression patterns among teleost fish, with tambaqui exhibiting a unique profile consistent with its migratory lifestyle and varied diet. Conclusions: This study offers new insights into the evolution and functional diversification of lipoprotein lipases in vertebrates, highlighting the complexity of lipid metabolism in fish. These findings contribute to understanding the adaptability of teleost fish to diverse environments and lay the foundation for future research in lipid metabolism regulation, including Neotropical species, with potential applications in aquaculture and conservation.
Full article
(This article belongs to the Special Issue Molecular Genetics Applied to Aquaculture: From Breeding Stock Selection to Biotechnological Innovations)
►▼
Show Figures
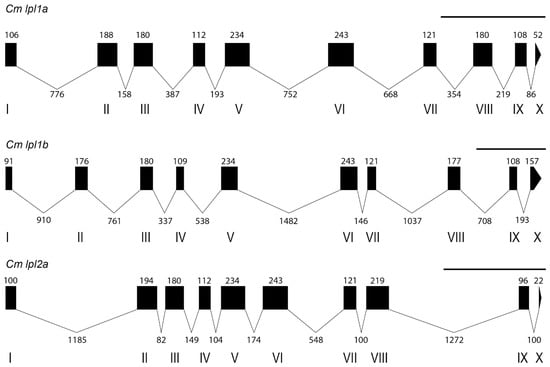
Figure 1

Journal Menu
► ▼ Journal Menu-
- Genes Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Diversity, Forests, Genes, IJPB, Plants
Plant Chloroplast Genome and Evolution
Topic Editors: Chao Shi, Lassaâd Belbahri, Shuo WangDeadline: 31 August 2025
Topic in
Applied Sciences, BioMedInformatics, BioTech, Genes, Computation
Computational Intelligence and Bioinformatics (CIB)
Topic Editors: Marco Mesiti, Giorgio Valentini, Elena Casiraghi, Tiffany J. CallahanDeadline: 30 September 2025
Topic in
Agriculture, Agronomy, Crops, Genes, Plants, DNA
Vegetable Breeding, Genetics and Genomics, 2nd Volume
Topic Editors: Umesh K. Reddy, Padma Nimmakayala, Georgia NtatsiDeadline: 31 October 2025
Topic in
Brain Sciences, CIMB, Epigenomes, Genes, IJMS, DNA
Genetics and Epigenetics of Substance Use Disorders
Topic Editors: Aleksandra Suchanecka, Anna Maria Grzywacz, Kszysztof ChmielowiecDeadline: 15 November 2025

Conferences
Special Issues
Special Issue in
Genes
Study on Genotypes and Phenotypes of Neurodegenerative Diseases—2nd Edition
Guest Editor: Claudia RicciDeadline: 15 May 2025
Special Issue in
Genes
Genome and Molecular Biology of Viruses
Guest Editors: Jun Luo, Xiaofeng GuoDeadline: 20 May 2025
Special Issue in
Genes
Abiotic Stress in Plants: Molecular Genetics and Genomics—2nd Edition
Guest Editor: Fei GaoDeadline: 20 May 2025
Special Issue in
Genes
Genetics and Genomics of Lung Cancer
Guest Editor: Elisa FrullantiDeadline: 20 May 2025
Topical Collections
Topical Collection in
Genes
Feature Papers in ‘Animal Genetics and Genomics’
Collection Editors: Antonio Figueras, Raquel Vasconcelos
Topical Collection in
Genes
Feature Papers: 'Plant Genetics and Genomics' Section
Collection Editors: Bin Yu, Roberto Tuberosa, Jacqueline Batley





